Data profiling in Python
Data profiling is intended to help understand data leading to a better data prepping and data quality.
Data profiling is the systematic up front analysis of the content of a data source, all the way from counting the bytes and checking cardinalities up to the most thoughtful diagnosis of whether the data can meet the high level goals of the data warehouse. Ralph Kimball
pandas-profiling Python package is a great tool to create HTML profiling reports. For a given dataset it computes the following statistics:
- Essentials: type, unique values, missing values
- Quantile statistics like minimum value, Q1, median, Q3, maximum, range, interquartile range
- Descriptive statistics like mean, mode, standard deviation, sum, median absolute deviation, coefficient of variation, kurtosis, skewness
- Most frequent values
- Histogram
- Correlations highlighting of highly correlated variables, Spearman and Pearson matrixes
Prerequisites
- Install Python and the Jupyter Notebook
- Install pandas-profiling module
- GitHub repo: https://github.com/pandas-profiling/pandas-profiling
- With Anaconda:
conda install -c conda-forge pandas-profiling
Profiling data from a CSV file
Let’s see how to use pandas-profiling package on a dataset from the University of California, Irvine: Communities and Crime Data Set
Start Jupyter Notebook
jupyter notebook
Create a new notebook
Load pandas and pandas_profiling
import pandas as pd
import pandas_profiling
Load data into a DataFrame object and cast categorical numeric columns
df=pd.read_csv("./data/communities.csv" , sep=',', na_values=["?"])
# Some other useful read_csv parameters
# encoding='UTF-8'
# low_memory=False
# parse_dates=["date_column_name"]
# Change Pandas DataFrame column data type
# df["y"] = pd.to_numeric(df["y"])
# df["z"] = pd.to_datetime(df["z"])
df['state'] = df['state'].astype('category')
df['community'] = df['community'].astype('category')
df['fold'] = df['fold'].astype('category')
Display the data profiling report in a Jupyter notebook
pandas_profiling.ProfileReport(df)
Or export it in an HTML file
profile = pandas_profiling.ProfileReport(df)
profile.to_file(outputfile="./report.html")
Profiling data from SQL Server database
## From SQL to DataFrame Pandas
import pandas as pd
import pyodbc
sql_conn = pyodbc.connect('DRIVER={SQL Server Native Client 11.0}; \
SERVER=server_name; \
DATABASE=db_name; \
Trusted_Connection=yes')
query = "SELECT [BusinessEntityID],[FirstName],[LastName],
[PostalCode],[City] FROM [Sales].[vSalesPerson]"
df = pd.read_sql(query, sql_conn)
pandas_profiling.ProfileReport(df)
With credentials replace pyodbc.connect instruction with the following
sql_conn = pyodbc.connect('DRIVER={SQL Server Native Client 11.0}; \
SERVER=server_name; \
DATABASE=db_name; \
uid=User; \
pwd=password')
As alternative you could consider the pymssql module.
Other data profiling commands
df.head()
df.describe()
df.isnull().any()
df.isnull().sum()
null_columns=df.columns[df.isnull().any()]
df[null_columns].isnull().sum()
df.isnull().any().any()
df.info()
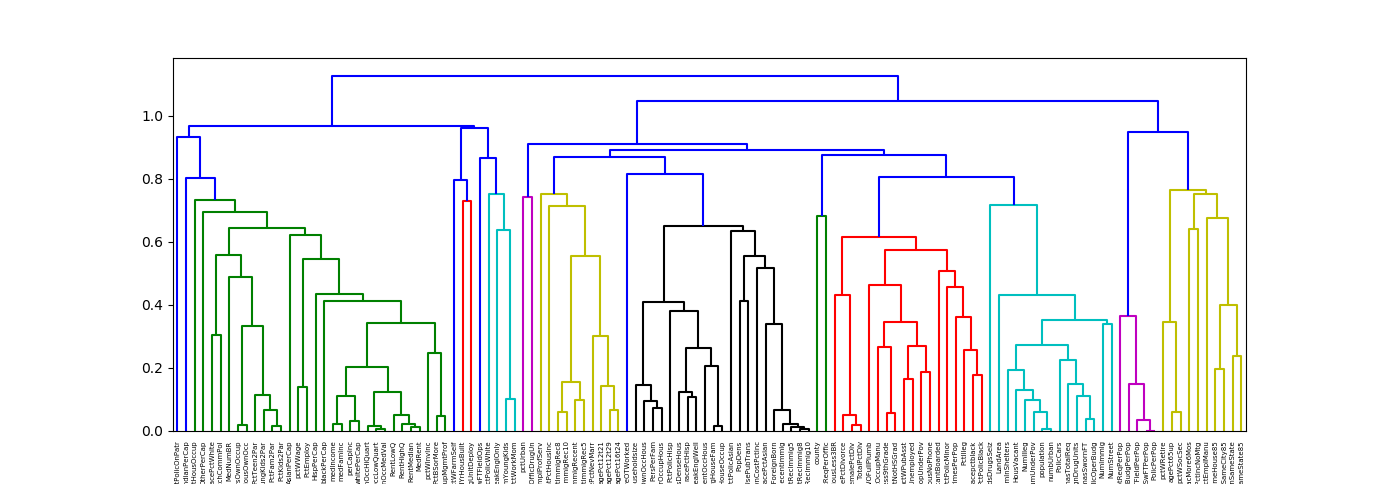
Plot a hierarchical clustering as a dendrogram.
%matplotlib notebook
import matplotlib.pyplot as plt
from scipy.cluster import hierarchy as hc
corr = 1 - df.corr()
corr_condensed = hc.distance.squareform(corr) # convert to condensed
z = hc.linkage(corr_condensed, method='average')
dendrogram = hc.dendrogram(z, labels=corr.columns)
Principal Component Analysis (PCA) in Python (https://stackoverflow.com/a/50572561/9489744)
%matplotlib notebook
import numpy as np
import matplotlib.pyplot as plt
from sklearn import datasets
from sklearn.decomposition import PCA
import pandas as pd
from sklearn.preprocessing import StandardScaler
iris = datasets.load_iris()
X = iris.data
y = iris.target
X = iris.data
y = iris.target
#In general a good idea is to scale the data
scaler = StandardScaler()
scaler.fit(X)
X=scaler.transform(X)
pca = PCA()
x_new = pca.fit_transform(X)
def myplot(score,coeff,labels=None):
xs = score[:,0]
ys = score[:,1]
n = coeff.shape[0]
scalex = 1.0/(xs.max() - xs.min())
scaley = 1.0/(ys.max() - ys.min())
plt.scatter(xs * scalex,ys * scaley, c = y)
for i in range(n):
plt.arrow(0, 0, coeff[i,0], coeff[i,1],color = 'r',alpha = 0.5)
if labels is None:
plt.text(coeff[i,0]* 1.15, coeff[i,1] * 1.15, "Var"+str(i+1), color = 'g', ha = 'center', va = 'center')
else:
plt.text(coeff[i,0]* 1.15, coeff[i,1] * 1.15, labels[i], color = 'g', ha = 'center', va = 'center')
plt.xlim(-1,1)
plt.ylim(-1,1)
plt.xlabel("PC{}".format(1))
plt.ylabel("PC{}".format(2))
plt.grid()
#Call the function. Use only the 2 PCs.
myplot(x_new[:,0:2],np.transpose(pca.components_[0:2, :]))
plt.show()

See also
date_range 02/09/2020
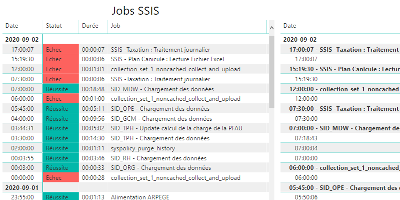
How to monitor SSIS job and package executions.
date_range 15/08/2020
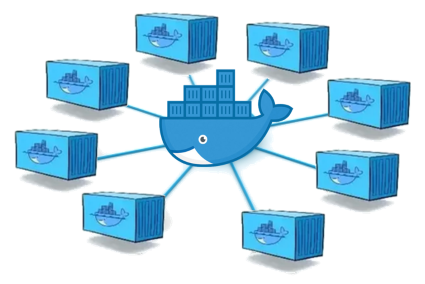
Enable a network connectivity between Docker containers on CentOS 8.
date_range 07/04/2020
Sphinx and GitHub provide an efficient and free way to publish your documentation online. Here we describe how to do so.